Day 4 of dashboard week dawned and we were promised easier prep… Seemingly this was the case upon first look. Andy had split us by different food types and tasked us with drawing the data from USDA Food Composition Databases – check for more details of the project here. This website contains tables of nutritional information within each entry of the database as well as an API (limited to 1000 calls per day). Apprehensive of taking the API approach – only to be chucked out halfway through, we decided to take the old-fashioned scrapping route. This proved great practice and a learning experience for me as I hadn’t done much before. Therefore, in this blog, I’ll focus on the data scrapping aspect of the project and share with you the method used to complete this task.
After searching for my food group (pizza) the database provided a table of results, where by clicking the link on each field, a page would open with more info and the option to download a .csv file.
The first step was to identify the right portion of HTML that contains all the desired links. By right clicking on the results page, clicking ‘inspect’, using the select tool (top left of the window next to mobile view) and finding the table body or ‘tbody’ (highlighted both on web page and in inspect).
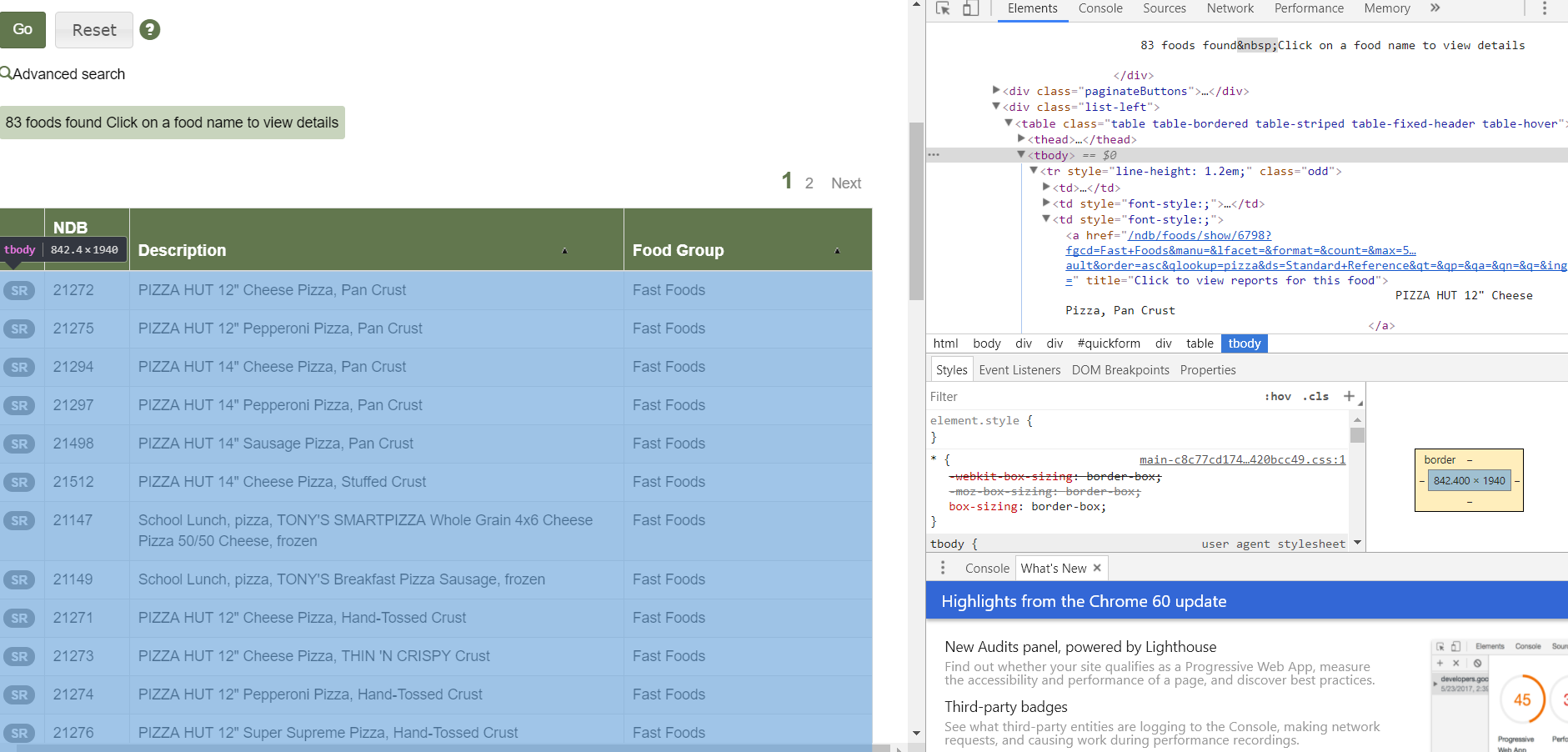
Next, right-click on ‘tbody’ and copy element:
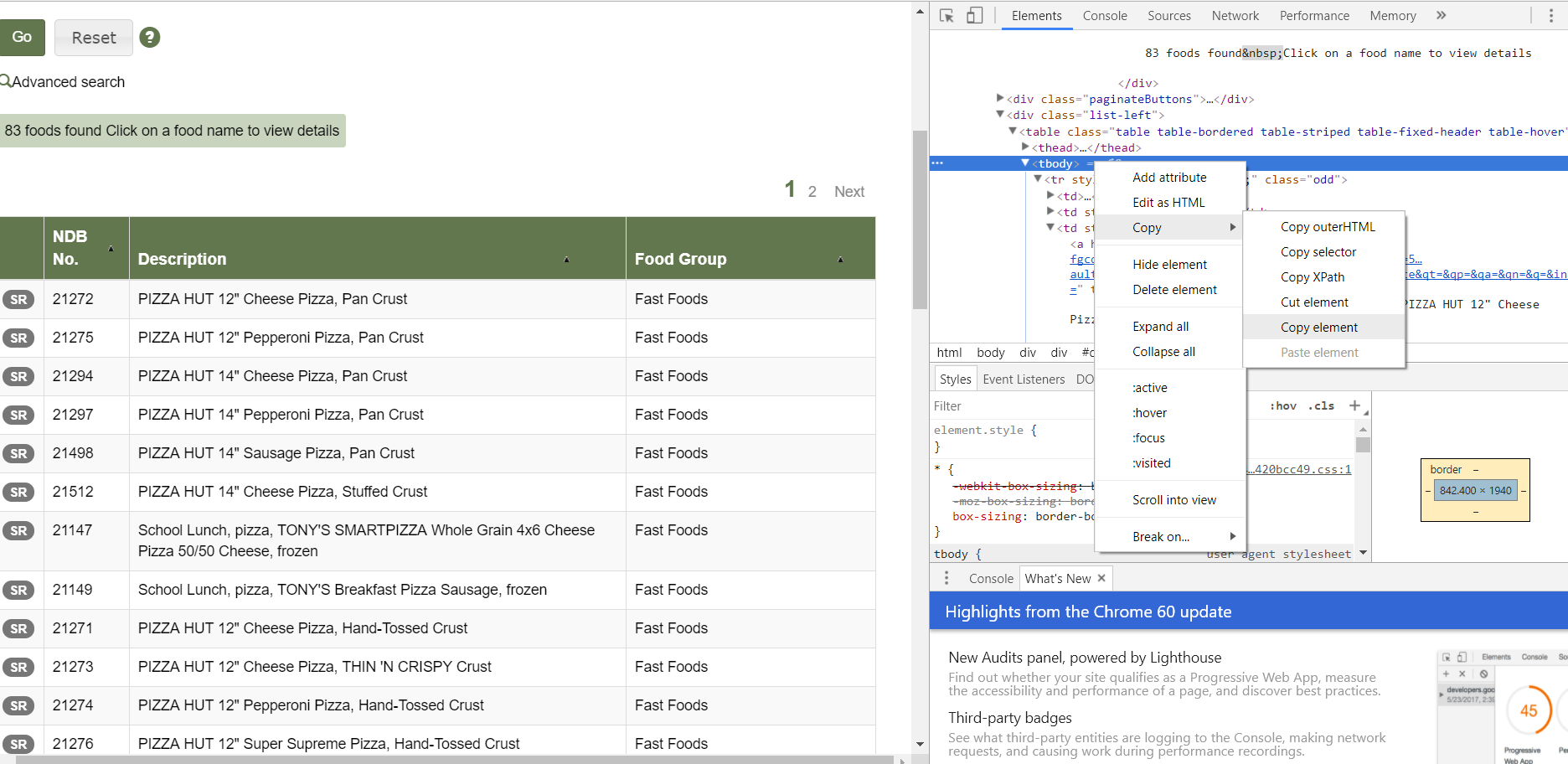
Paste into a text file:
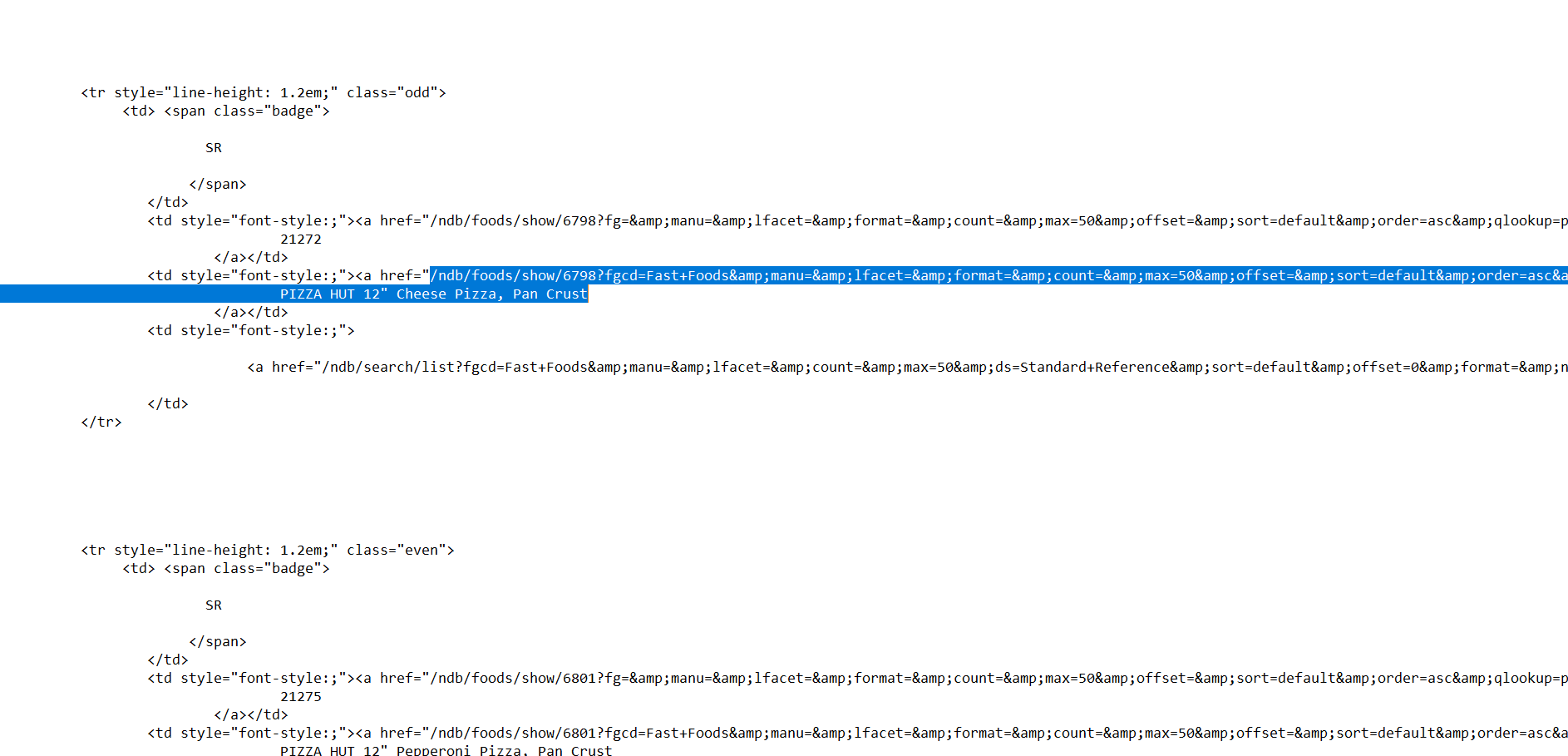
Within this mumbo jumbo are the links to each individual entry, I have highlighted the first in the picture above. Notice all the links have ‘href=’ before the link, this can be used to filtering when in Alteryx so that only links are obtained. There are however 3 links per entry and so further filtering for something individual about the specific entry link we want like ‘fgcd’ can be used to single it out.
So, just to be clear on the objective of this exercise, in this instance by taking:
The base URL ‘ndb.nal.usda.gov/’
+
the individual food entry part of the link i.e.: ‘ndb/foods/show/6798?’
+
the final part to trigger the .csv download
Consequently, we can generate the download link for the .csv download file for each entry in the results.
The .csv trigger part is highlighted in the text file below and is retrieved by copying the download link:
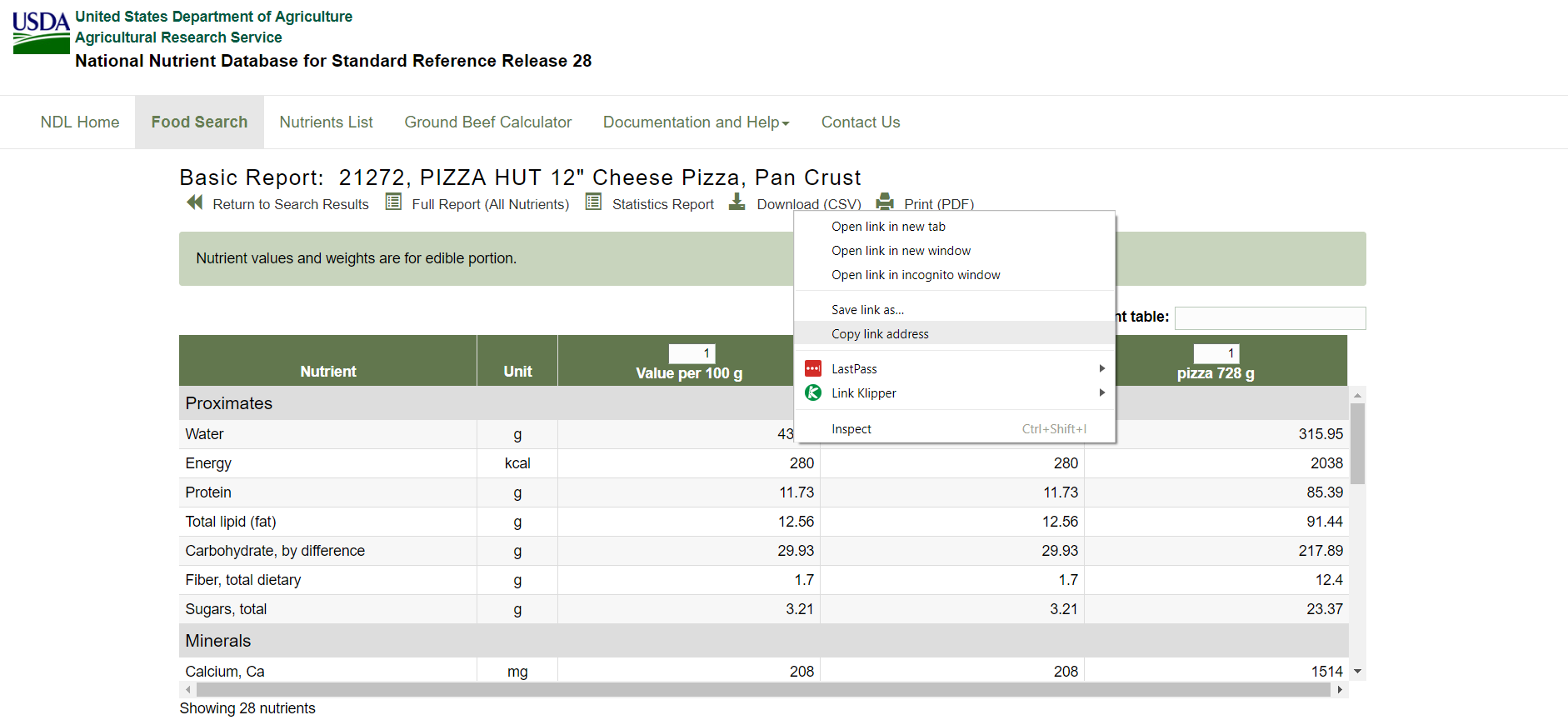
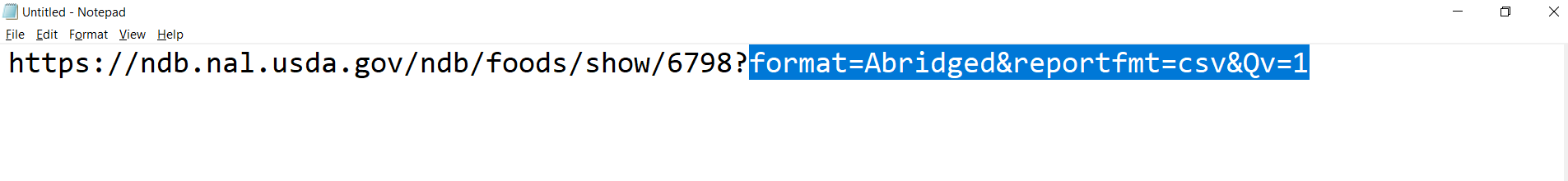
The whole link in the text file above is an example of one of the desired links to be produced on mass. Doing this all in Alteryx means that a considerable number of downloads (the whole tbody of links) can be achieved with just one or two workflows:
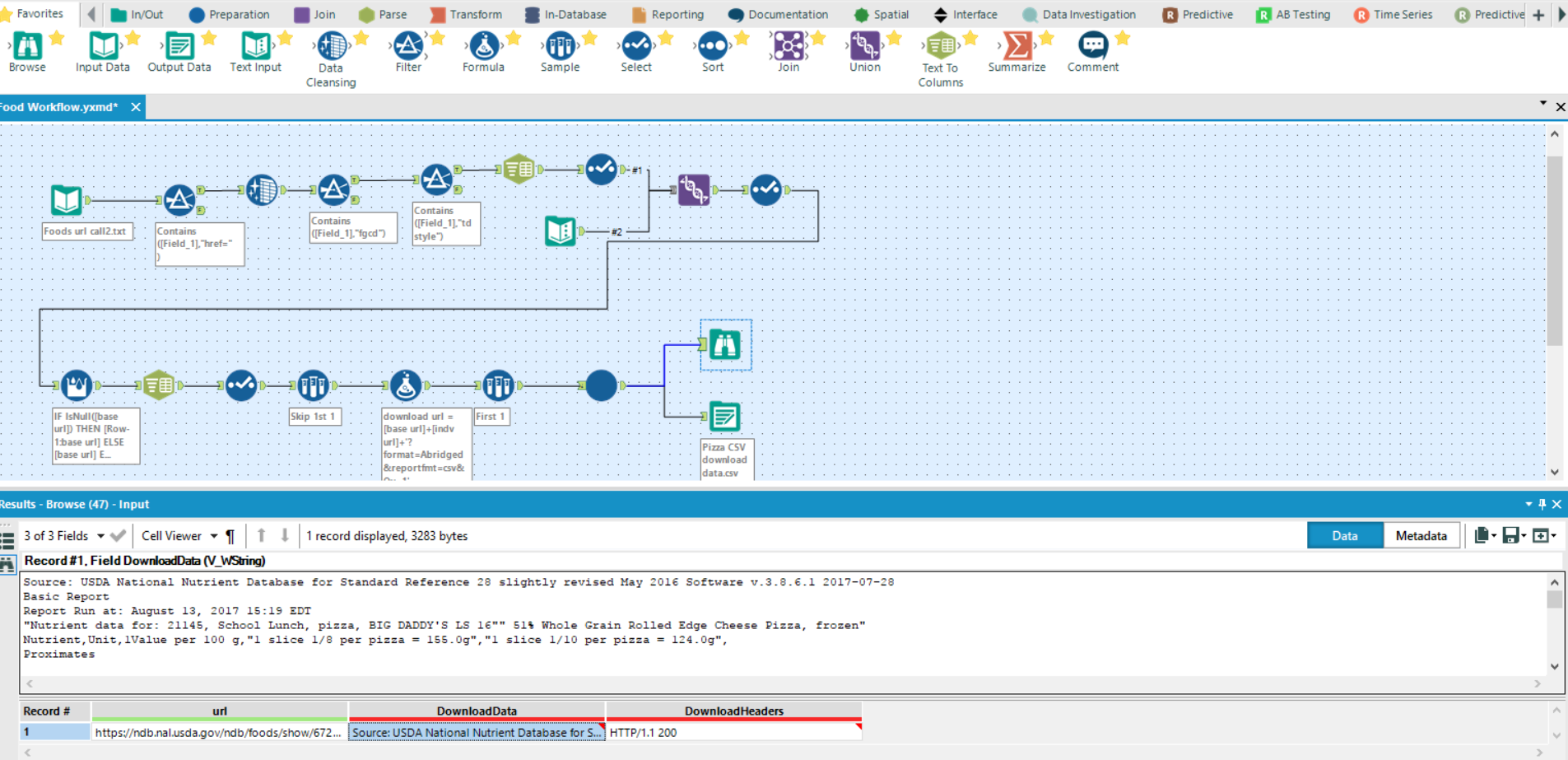
This workflow is just doing the previously described: filtering for desired link parts (individual entry identifier), adding it after the base URL (text input at union), adding in the link command for .csv download, then running through a crew macro called ‘wait a sec’ to stagger the download calls as too many at once would return errors (check Will’s blog for more info on this batch macro).
N.B. – you can download tools such as ParseHub or Link Klipper that will pull links from pages as well, which are almost definitely quicker, however, I wanted to practice the method described.
Next, It was a case of unlocking the puzzle of information that was the .csv file itself, in a long, arduous clean and prep workflow to put the data into a usable structure:
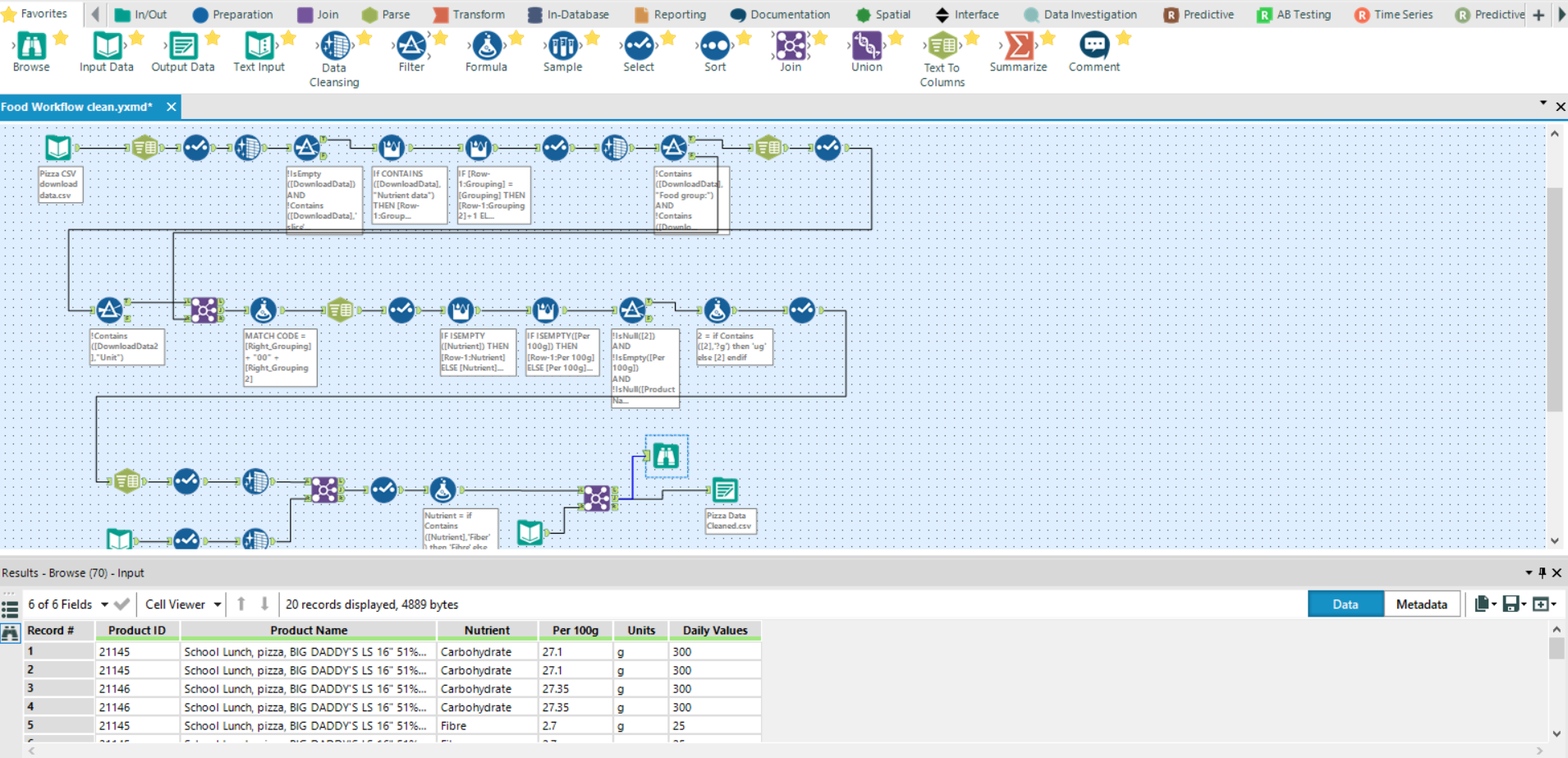
The structure of the .csv file will differ between sources; therefore, some will require more clean-up than others. Here, as a basic overview, I have: imported the download data obtained from the first workflow, split it into rows, filtered for relevant info, built record ID’s for entries, split the product names and re-joined back onto the data, filtered again for relevant info and then added in additional recommended daily intake values.
Over to Tableau and I wanted to create a dashboard that used the food packaging concept to identify low, medium or high levels of nutrients in food.
See in the picture below or interact here:
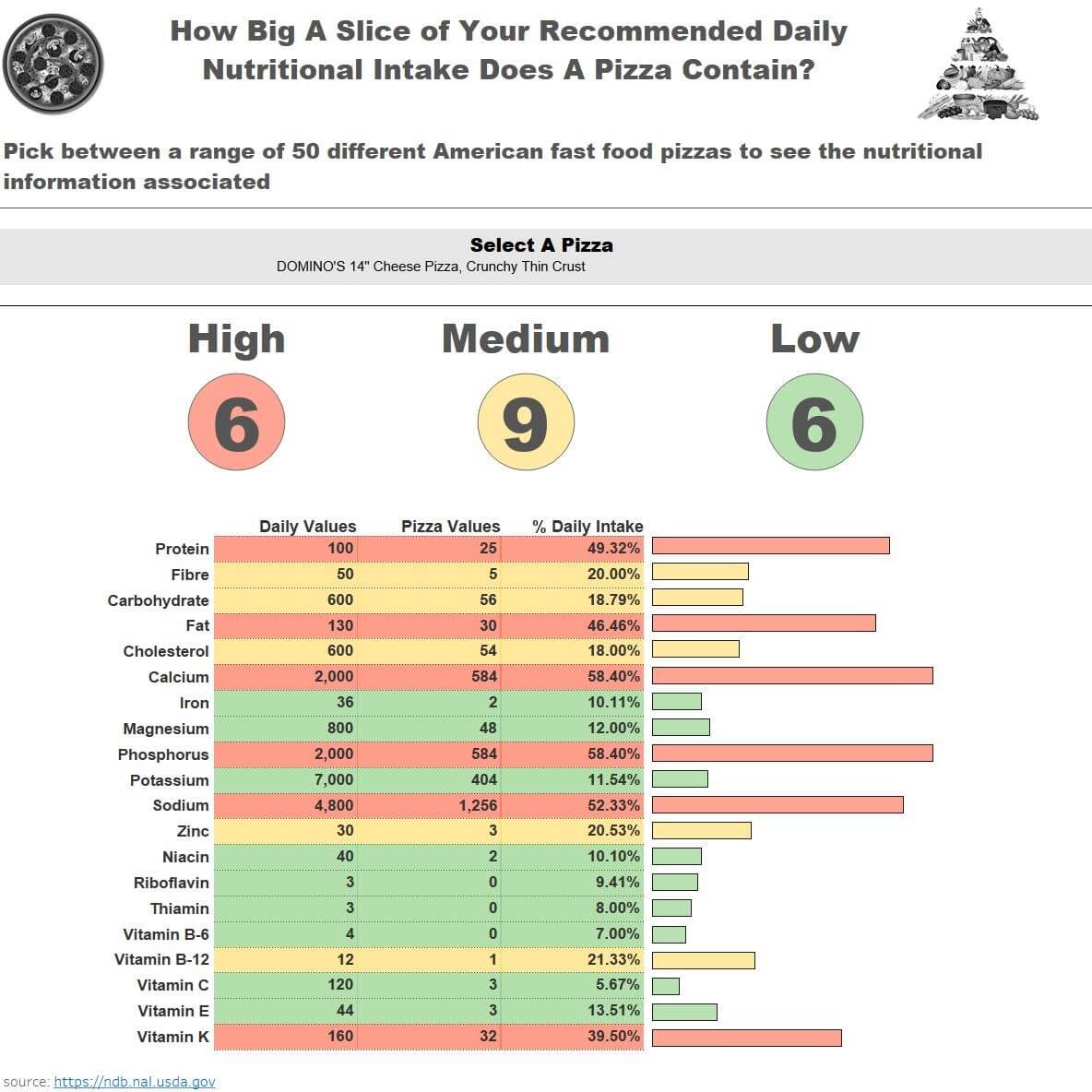
I realise however more work is required as for some foods it is good to have a high level of nutrients and for others, it is not so. As a result, I would either restructure, separate into two groups or add further calculations to cater for this.
